Pedigree analysis of one familial hypertrophic cardiomyopathy caused by rare homozygous mutation of TNNI3(p. Arg79Cys) and bioinformatics analysis of the mutation
-
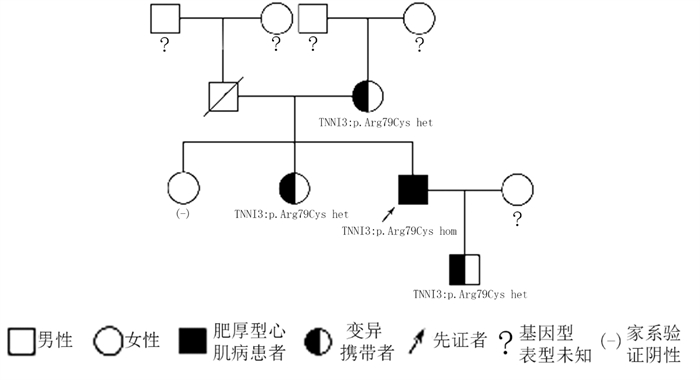
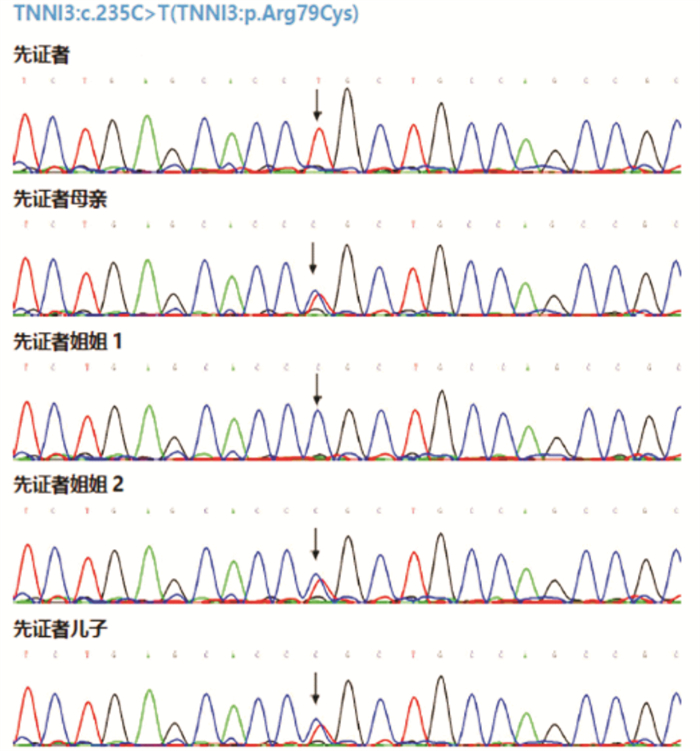
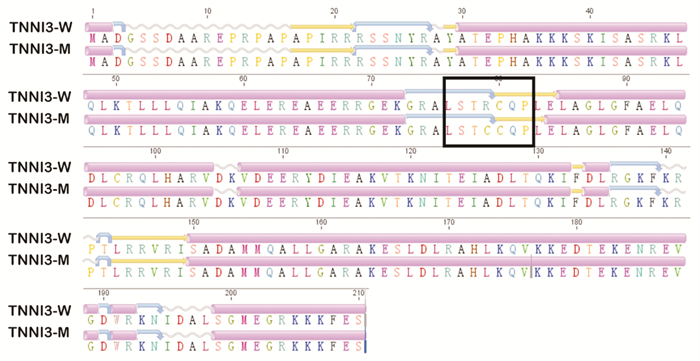
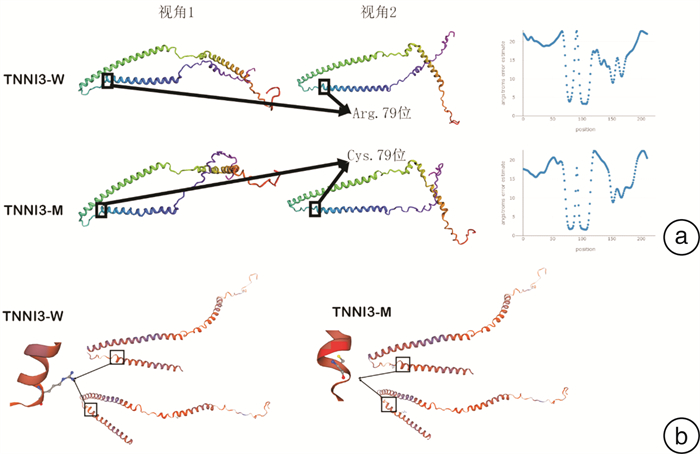
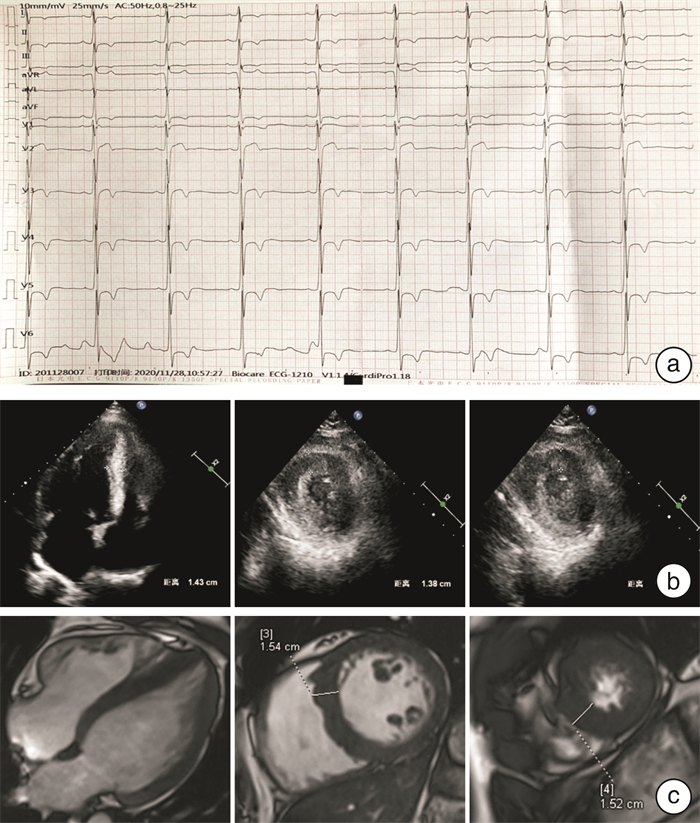
摘要: 目的 对TNNI3基因p.Arg79Cys罕见纯合突变导致一家族性肥厚型心肌病进行家系分析,并对突变位点进行生物信息学分析。方法 对先证者及其亲属进行临床资料的评估,采集先证者及其亲属静脉血提取全基因组DNA并进行二代测序,寻找致病基因并进行Sanger测序验证。对致病基因突变进行生物信息学分析,获取突变基因野生型及突变型氨基酸序列,利用计算机对野生型及突变型蛋白质进行二级结构及三级结构预测分析。结果 先证者直系亲属均不符合肥厚型心肌病诊断标准。先证者TNNI3基因5号外显子p.Arg79Cys的纯合突变,先证者母亲、其中1个姐姐及儿子携带TNNI3基因杂合突变,另外1个姐姐未携带TNNI3基因突变。生物信息学分析提示该变异所致的氨基酸改变可能会对蛋白功能产生影响,并且导致蛋白质稳定性降低,在脊椎动物中保守性较好。二级结构预测突变的TNNI3及野生型TNNI3蛋白质二级结构未见明显变化。三级结构预测蛋白三级结构转角处折叠的角度发生了变化,导致蛋白的空间结构发生了变化。结论 TNNI3基因p.Arg79Cys纯合突变可能通过不同于经典单基因遗传的其他遗传模式导致肥厚型心肌病,该位置的突变对蛋白质的结构产生了影响。Abstract: Objective The study aims to perform pedigree analysisof one familial hypertrophic cardiomyopathy(HCM) caused by a rare homozygous mutation ofTNNI3, as well as carry out bioinformatics analysis of the mutation.Methods The clinical data of the proband and his relatives were evaluated. Venous blood was collected from the proband and his family. The whole genome DNA was extracted and screened by targeted next-generation sequencing, following validation by Sanger sequencing. Bioinformatics analysis of the mutation site was carried out. The amino acid sequences of wild-type and mutantTNNI3 were obtained, and the secondary and tertiary structures of the proteins were predicted and analyzed by a computer.Results None of the proband's family members met the diagnostic criteria of HCM. A homozygous mutation ofTNNI3 P. Arg79Cys was found in the proband. The proband's mother, one sister and son carried the heterozygous mutation of the same site while the other sister did not carry the mutation. Sequence alignment showed the residue homology in closely related mammals. Bioinformatics analysis indicated that the changes of the amino acid caused by this mutation might affect the protein function. In addition, the stability of mutant protein was decreased. There was no significant change in the secondary structure between the wild-type TNNI3 and the mutant protein. It was predicted that the angle at the corner of the tertiary structure changed, resulting in the change of the spatial structure of the protein.Conclusion The homozygous mutation ofTNNI3 P. arg79cys may lead to hypertrophic cardiomyopathy through other genetic modes different from the classical single gene inheritance. The mutation at this site has an impact on the structure of the protein.
-
Key words:
- hypertrophic cardiomyopathy /
- TNNI3 /
- homozygous mutation /
- protein structure
-
-
-
[1] 林丽容, 卢荔红, 胡雪群, 等. 家族性肥厚型心肌病MYBPC3基因变异及其临床表型分析[J]. 临床心血管病杂志, 2021, 37(6): 557-560. https://www.cnki.com.cn/Article/CJFDTOTAL-LCXB202106013.htm
[2] Salazar-Mendiguchía J, Ochoa JP, Palomino-Doza J, et al. Mutations in TRIM63 cause an autosomal-recessive form of hypertrophic cardiomyopathy[J]. Heart, 2020, 106(17): 1342-1348. doi: 10.1136/heartjnl-2020-316913
[3] Dorsch LM, Schuldt M, dos Remedios CG, et al. Protein Quality Control Activation and Microtubule Remodeling in Hypertrophic Cardiomyopathy[J]. Cells, 2019, 8(7).
[4] 段丽琴, 李琼, 任毅, 等. TNNI3基因p. Arg162Gln罕见纯合突变所致肥厚型心肌病一家系[J]. 中华心血管病杂志, 2019, 47(12): 1008-1010. doi: 10.3760/cma.j.issn.0253-3758.2019.12.012
[5] Pantou MP, Gourzi P, Gkouziouta A, et al. A case report of recessive restrictive cardiomyopathy caused by a novel mutation in cardiac troponin I(TNNI3)[J]. BMC Med Genet, 2019, 20(1): 61. doi: 10.1186/s12881-019-0793-z
[6] Tao Q, Yang J, Cheng W, et al. A Novel TNNI3 Gene Mutation(c. 235C>T/p. Arg79Cys)Found in a Thirty-eight-year-old Women with Hypertrophic Cardiomyopathy[J]. Open Life Sci, 2018, 13: 374-378. doi: 10.1515/biol-2018-0045
[7] Ciabatti M, Fumagalli C, Beltrami M, et al. Prevalence, causes and predictors of cardiovascular hospitalization in patients with hypertrophic cardiomyopathy[J]. Int J Cardiol, 2020, 318: 94-100. doi: 10.1016/j.ijcard.2020.07.036
[8] Yang Q, Wang B, Wang J, et al. Analysis of genotype-phenotype correlation for a novel MYH7-D554Y mutation identified in an ethnic Han Chinese pedigree affected with hypertrophic cardiomyopathy[J]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 2018, 35(5): 667-671. http://www.ncbi.nlm.nih.gov/pubmed/30298491
[9] Mathur MC, Kobayashi T, Chalovich JM. Some cardiomyopathy-causing troponin I mutations stabilize a functional intermediate actin state[J]. Biophys J, 2009, 96(6): 2237-2244. doi: 10.1016/j.bpj.2008.12.3909
[10] Doevendans PA, Glijnis PC, Kranias EG. Leducq Transatlantic Network of Excellence to Cure Phospholamban-Induced Cardiomyopathy(CURE-PLaN)[J]. Circ Res, 2019, 125(7): 720-724. doi: 10.1161/CIRCRESAHA.119.315077
[11] Hammer E, Goritzka M, Ameling S, et al. Characterization of the human myocardial proteome in inflammatory dilated cardiomyopathy by label-free quantitative shotgun proteomics of heart biopsies[J]. J Proteome Res, 2011, 10(5): 2161-2171. doi: 10.1021/pr1008042
[12] Yu H, He X, Liu X, et al. A novel missense variant in cathepsin C gene leads to PLS in a Chinese patient: A case report and literature review[J]. Mol Genet Genomic Med, 2021, 9(7): e1686. http://onlinelibrary.wiley.com/doi/pdf/10.1002/mgg3.1686
[13] Maron BJ, Rowin EJ, Maron MS. Paradigm of Sudden Death Prevention in Hypertrophic Cardiomyopathy[J]. Circ Res, 2019, 125(4): 370-378. doi: 10.1161/CIRCRESAHA.119.315159
[14] Husser D, Ueberham L, Jacob J, et al. Prevalence of clinically apparent hypertrophic cardiomyopathy in Germany-An analysis of over 5 million patients[J]. PLoS One, 2018, 13(5): e0196612. doi: 10.1371/journal.pone.0196612
[15] Teekakirikul P, Zhu W, Huang HC, et al. Hypertrophic Cardiomyopathy: An Overview of Genetics and Management[J]. Biomolecules, 2019, 9(12).
[16] Wolf CM. Hypertrophic cardiomyopathy: genetics and clinical perspectives[J]. Cardiovasc Diagn Ther, 2019, 9(Suppl 2): S388-S415.
[17] Kitaoka H, Kubo T, DoiYL. Hypertrophic Cardiomyopathy-A Heterogeneous and Lifelong Disease in the Real World[J]. Circ J, 2020, 84(8): 1218-1226. doi: 10.1253/circj.CJ-20-0524
[18] Guerrier K, Anderson JB, Pratt J, et al. Correlation of precordial voltages to left ventricular mass on echocardiogram in adolescent patients with hypertrophic cardiomyopathy compared with that in adolescent athletes[J]. Am J Cardiol, 2015, 115(7): 956-961. doi: 10.1016/j.amjcard.2015.01.025
[19] Kimura A, Harada H, Park JE, et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy[J]. Nat Genet, 1997, 16(4): 379-382. doi: 10.1038/ng0897-379
[20] Mogensen J, Murphy RT, Kubo T, et al. Frequency and clinical expression of cardiac troponin I mutations in 748 consecutive families with hypertrophic cardiomyopathy[J]. J Am Coll Cardiol, 2004, 44(12): 2315-2325. doi: 10.1016/j.jacc.2004.05.088
[21] Wu G, Liu L, Zhou Z, et al. East Asian-Specific Common Variant in TNNI3 Predisposes to Hypertrophic Cardiomyopathy[J]. Circulation, 2020, 142(21): 2086-2089. doi: 10.1161/CIRCULATIONAHA.120.050384
[22] Gurtner C, Hug P, Kleiter M, et al. YARS2 Missense Variant in Belgian Shepherd Dogs with Cardiomyopathy and Juvenile Mortality[J]. Genes(Basel), 2020, 11(3).
-



 下载:
下载: